![]()
![]()
In the previous investigation (Powering Life) it was noted that energy is never created or destroyed, but captured by life and stored in the chemical bonds in organic molecules. Plants package captured sunlight energy mostly in carbohydrate molecules. Animals eat such chemical food then extract the energy through a series of small, often reversible steps into a universal energy supplying molecule, adenosine triphoshate, better known by its initials, ATP. The ATP is used whenever energy is needed to build and repair the organism, and to operate life processes.
But the life machinery that captures and uses the energy in chemical form is itself an assembly of chemical molecules. How does such a living machine create and assemble itself? The control of the process is done by DNA and RNA, specialized chain molecules composed of nucleic acid subunits. DNA and RNA direct protein molecules to both serve as factories assembling other molecules and as enzymes to control all biochemical reactions. This process is done in two stages: The first stage in or near mitochondria produces a pool of relatively small molecules which serve as building blocks for the specialized larger molecules. The second stage assembles the building blocks into useful large molecules under the genetic control of the cell nucleus. The reproduction of the coded genetic information itself will be outlined in the next investigation, (Reproducing Life).
The previous investigation noted that since humans are omnivores eating a wide variety of different foods, our bodies can metabolize the diverse food molecules, carbohydrates, lipids, proteins and nucleic acids. Each is digested to small molecules for absorption into the body and transported to individual cells where they are metabolized to even simpler molecules for conversion in the Krebs cycle inside mitochondria to CO2 with their Hydrogen transported away by NAD and FAD. The Hydrogen and associated electrons are delivered to a chain of proteins embedded in the inner mitochondria membrane where ATP is assembled from the captured energy by a process which temporarily stores the energy in a pumped up concentration of Hydrogen ions. In the final step, Hydrogen ions and electrons are combined with corrosive Oxygen to form H2O.
But not all molecules end as CO2 and H2O. Useful intermediates are drawn off mid-process. Because many of the reactions are reversible, and alternate enzymes can often substitute for remaining reactions, the Krebs Cycle provides feed-stock for producing the lipids, proteins, and nucleic acids and all the other molecules needed to assemble and repair a living organism.
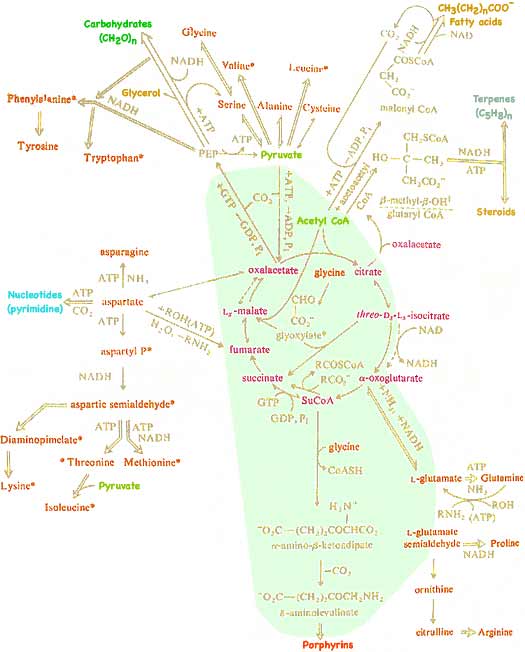
The reactions in the light green area occur inside mitochondria. Those in the white region occur nearby within cells. It should now be clear that while the Kreb Cycle is the central mechanism in extracting energy from food, it is also universally used by living organisms to transform available molecules into molecular building units needed to make the large, functional molecules essential for an organism's growth and repair. You can eat one kind of molecule yet your body can transform that to a different needed molecule.
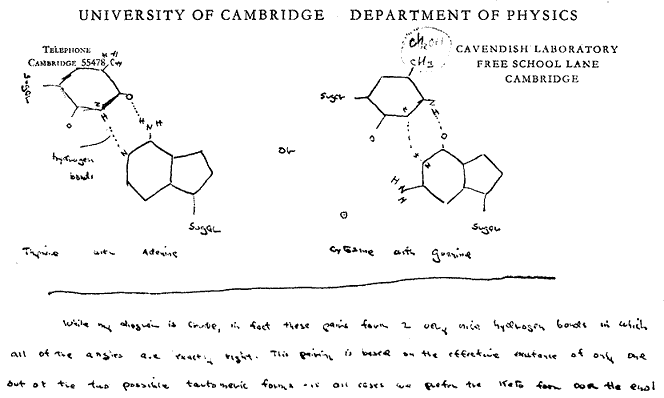
 During 1868 Friedrich Miescher, a Swiss physician, isolated a new type of compound from cell nuclei which we now call nucleic acid. That discovery was only published in 1890 after his death. In 1944 O.T. Avery in New York succeeded in transferring a heritable property from one bacterium to another using pure deoxyribonucleic acid, demonstrating that genetic information, previously thought due to proteins, is actually carried by nucleic acids. Abbreviated as DNA, this is a giant polymer chain alternates a phosphate group with the sugar deoxyribose attached to with one of four nitrogen-containing chemical bases. Maurice Wilkins (b1916, d2004), who earlier suspected the importance of DNA had X-rays of deoxyribonucleic acid made which showed that DNA was a twisted helix. Crystallographer, Rosalind Franklin (b1920, d1958), from whom Wilkins sought assistance, made superior X-ray diffraction images which she claimed revealed that the phosphate groups were on the outside of the helix. Emulating Linus Pauling, James Watson (b1928 far right) and Francis Crick (b1916, d2004 near right) in Cambridge tried to build models to fit her X-ray pattern and data. They concluded that the X-ray patterns showed that DNA is double strand then showed that a single-ring-base paired by hydrogen bonds with a double-ring-base exactly fit the distance between the two helix chains. They proposed that the uniquely bonded pairs of adenine with thymine, cytosine with guanine, and their two mirrored isomers (as shown in Watson's letter below) match the DNA helix dimensions, explained the genetic code, that DNA specified the amino acid sequences in proteins, and how the code could be reproduced in cell division. Their 1953 proposal for DNA led to the 1962 Nobel Prize in Physiology or Medicine being awarded to Wilkins, Watson, and Crick. Rosalind Franklin, perhaps slighted by gender bias as she was considered Wilkins assistant rather than equal, was not included because Nobels are only awarded living scientists.
During 1868 Friedrich Miescher, a Swiss physician, isolated a new type of compound from cell nuclei which we now call nucleic acid. That discovery was only published in 1890 after his death. In 1944 O.T. Avery in New York succeeded in transferring a heritable property from one bacterium to another using pure deoxyribonucleic acid, demonstrating that genetic information, previously thought due to proteins, is actually carried by nucleic acids. Abbreviated as DNA, this is a giant polymer chain alternates a phosphate group with the sugar deoxyribose attached to with one of four nitrogen-containing chemical bases. Maurice Wilkins (b1916, d2004), who earlier suspected the importance of DNA had X-rays of deoxyribonucleic acid made which showed that DNA was a twisted helix. Crystallographer, Rosalind Franklin (b1920, d1958), from whom Wilkins sought assistance, made superior X-ray diffraction images which she claimed revealed that the phosphate groups were on the outside of the helix. Emulating Linus Pauling, James Watson (b1928 far right) and Francis Crick (b1916, d2004 near right) in Cambridge tried to build models to fit her X-ray pattern and data. They concluded that the X-ray patterns showed that DNA is double strand then showed that a single-ring-base paired by hydrogen bonds with a double-ring-base exactly fit the distance between the two helix chains. They proposed that the uniquely bonded pairs of adenine with thymine, cytosine with guanine, and their two mirrored isomers (as shown in Watson's letter below) match the DNA helix dimensions, explained the genetic code, that DNA specified the amino acid sequences in proteins, and how the code could be reproduced in cell division. Their 1953 proposal for DNA led to the 1962 Nobel Prize in Physiology or Medicine being awarded to Wilkins, Watson, and Crick. Rosalind Franklin, perhaps slighted by gender bias as she was considered Wilkins assistant rather than equal, was not included because Nobels are only awarded living scientists.
With a pool of small building block amino acids provided by mitochondria and the surrounding cytoplasm, and the information provided by the DNA code, the cell assembles large proteins in ribosomes to serve as structural materials and to act as catalytic enzymes, controlling the biochemical reactions at the core of life. Ribosomes are a type of organelle made primarily of proteins and another nucleic acid, ribose nucleic acid. RNA is very similar to DNA except its sugar, ribose, has one additional Oxygen atom compared to DNA and uracil, one of the RNA bases, is missing a side methyl group present in DNA's thymine. Understanding the process used to assemble proteins started in the 1950s when F.C. Zamecnick, P. Siekevitz and later R. Schweet and M. Lamborg investigated functioning of ribosomes. Hoagland in Paul Zamecnik's laboratory at the Massachusetts General Hospital made two important discoveries. He found that amino acids are initially activated by ATP then transferred to small, water soluble RNA molecules (now known as transfer RNA = sRNA = tRNA), as an intermediate in formation of proteins. While obvious that amino acid activation would have to occur, Hoagland's second discovery (in 1956) of a previously undiscovered form of RNA (tRNA) was unanticipated by almost everybody. When previous attempts to determine how ribosomes could pick specific amino acids to assemble into a protein, Crick speculated an adapter molecule might be required. Hoagland and Paul Berg, established that the 20 or more tRNA molecules are in fact each specific for a particular amino acid.
Then it was found in 1959 that addition of DNA actually reduced the synthesis of protein! This suggested that a form of RNA, rather than DNA, might be the actual template for protein synthesis. From virus infected cells it was determined that its RNA attaches onto the host cell's ribosomes. Marshall W. Nirenberg (b 1927) convincingly confirmed the concept of messenger-RNA June 1961 by synthesizing a messenger RNA chain composed of only a single repeating RNA nucleotide, uracil. He demonstrated that ribosomes then use this mRNA code to produced a protein contained only the single amino acid, phenylalanine and that tRNA for phenylalanine was a required intermediate. Robert W. Holley (b1922, d1993) determined in 1965 the chemical structure of transfer-RNA which must provide the specific amino acid to match the code carried by mRNA. Har Gobind Khorana (b1922 in India) synthesized over about 6 years all 64 possible combinations of nucleotides to decipher the DNA code. As a result the 1968 Nobel Prize in Physiology or Medicine was awarded to these American scientists: Nirenberg, Holley and Khorana.
The synthesis of protein works in the following way:
start sequenceand begins tracking down the RNA.
The genetic code is a relatively simple language based on the equivalent of only 4 letters, the DNA bases adenine, cytosine, guanine, and thymine. Crick, Barnett, Brenner and Watts-Tobin confirmed that it carries information for the 20 amino acids by using exclusively 3 letter, non-overlapping words (which allow the 64 different combinations shown below). Khorana's determination of which amino acids were specified by which word was accomplished by synthesizing triples of nucleotides and analyzing which amino acids were selected. Eight amino acids only need the first two letter codes, leaving variations in the third letter as synonyms. Four of the words
serve as codons for starting or terminating synthesis of the protein.
| RNA
code for amino acids |
Second Nucleotide in word |
||||||||
| U | C | A | G | ||||||
| First Nucleotide |
U | UUU UUC |
phenyl- alanine |
UCU UCC UCA UCG |
serine | UAU UAC |
tyrosine | UGU UGC |
cysteine |
|---|---|---|---|---|---|---|---|---|---|
| UUA UUG |
leucine | UAA UAG |
STOP | UGA | STOP | ||||
| UGG | tryptohan | ||||||||
| C | CUU CUC CUA CUG |
leucine | CCU CCC CCA CCG |
proline | CAU CAC |
hisidine | CGU CGC CGA CGG |
arginine | |
| CAA CAG |
glutamine | ||||||||
| A | AUU AUC AUA |
isoleucine | ACU ACC ACA ACG |
threonine | AAU AAC |
asparagine | AGU AGC |
serine | |
| AAA AAG |
lysine | AGA AGG |
arginine | ||||||
| AUG | methionine START |
||||||||
| G | GUU GUC GUA GUG |
valine | GCU GCC GCA GCG |
alanine | GAU GAC |
aspartic acid |
GGU GGC GGA GGG |
glycine | |
| GAA GAG |
glutamic acid |
||||||||
The 3 dimensional shapes of protein are critical for their function and reactions. However once the amino acids are assembled in the correct sequence, random jostling by their environment's temperature is often sufficient to allow a protein to fold into a preferred shape, held there by its minimum potential energy.
Alternate shapes with nearly equal low potential energy allow many proteins to change shape and assume functions that a completely stiff shape would preclude. For example, a protein may function as an enzyme by temporarily binding onto anchor points on a substrate molecule, then changing its own shape to wrap around what is called the active site of the substrate to there catalyze a chemical reaction which would not have otherwise been likely. After the reaction has occurred, the flexible part of the enzyme might then move out of the way and allow the weak bonds to disengage so that the changed substrate can float free.
While there are several alternate codons (shown in above table) which code for most of the amino acids, it was long thought that as long as the amino acid sequence for a specific protein was the same, it made no difference which of the alternate nucleotide codes were used. However that is not always true. For at least some proteins, there are apparently multiple low potential energy shapes with significant energy barriers preventing easy conversion from one shape to another. Somehow alternate nucleotide codes result in forming the alternate shaped (but otherwise identically sequenced amino acids) proteins such that the two shape variations remain distinct and do not function identically. (The first example found was a protein, P-glycoprotein, which pumps chemotherapy drugs out of cells at two different rates depending on an apparent shape difference determined by which nucleotide code was used to manufacture the protein.)
Synthesis of protein from the DNA code is generally very accurate although errors are thousands of times more likely with some codons than others. Some junk code
segments of DNA do not produce protein, but appear to be due to be remnants left by virus infection. Such an infection mechanism is becoming more common as a way to patch and fix undesirable genetic code. For example cells which lack the nucleotide code needed to manufacture certain proteins can be infected with the code so that the cells gain the desired manufacturing capability.
 Many of the proteins serve as enzymes, organic catalysts which promote other molecules to assemble. As discussed using examples in previous investigations, often synthesis in living organisms uses a series of enzyme reactions using energy supplied by ATP when they would not otherwise be spontaneous.
Many of the proteins serve as enzymes, organic catalysts which promote other molecules to assemble. As discussed using examples in previous investigations, often synthesis in living organisms uses a series of enzyme reactions using energy supplied by ATP when they would not otherwise be spontaneous.
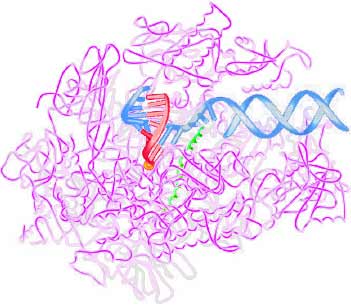
One of many key proteins is RNA polymerase which surrounds the (DNA shown as blue), separates the double strands, and oversees the assembly of the (red) messenger RNA. Roger Kornberg (b1947, below right) was awarded the 2006 Nobel Prize in Chemistry for pausing the transcription process (by withholding the next needed nucleotide), crystalizing the protein on a lipid membrane, and in 2001 using X-ray diffraction and electron microscopy to determine its structure (shown at right). This protein has a cavity,
just the right size so that it only accepts the RNA nucleotide which fits its matching DNA nucleotide. If the wrong RNA nucleotide tries to fit, just like a wrong jigsaw piece, it will not fit into the cavity. Once a new nucleotide has been inserted at the end of the RNA (with the help of a metal ion), the DNA-strand is ratcheted forward by a springy helical section.
Kornberg isolated another complex of proteins which provides an on-off switch for the transcription process. The DNA includes promoter
code segments, each of which with the help of the complex binds to specific substances found only in particular tissues. In this way a tissue containing a specific substance promotes the transcription process, makes the complimentary RNA needed to direct the assembly of protein needed by that tissue. Other tissues, lacking that particular substance, terminate the transcription of that section of DNA resulting in that available genetic code being ignored and none of the particular protein described by that genetic code being manufactured. Kornberg discovered that this regulation requires the presence of yet another molecular complex termed the Mediator, transmitting the signals and thereby switching the transcription on or off.
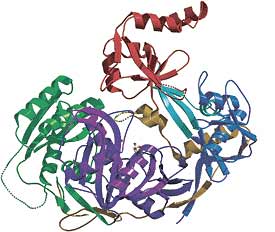 Small pieces of RNA also exercise control over gene expression by guiding the degradation of messenger RNA before it can be translated into protein by the cell's ribosomes. Several types of short RNAs provide powerful regulation of gene expression. The main classes of small RNAs are siRNAs (21 to 24 nucleotides long, so far identified in plant, fly, and worm cells, but also functional in mammalian cells), microRNAs (abbreviated miRNA), and PIWI-interacting, or piRNAs (24-31 nucleotides each, first isolated from reproductive cells of both flies and mannals), 21U RNAs (21 nucleotides long starting with uracil) and tncRNAs (noncoding RNAs from sections which don't code for proteins). These small RNAs are often cut off and separated from double-stranded RNA, the stem of an RNA hairpin which folds back on itself, or other sections of RNA which are not used as template for protein production. The small RNAs interacting with argonaute proteins (←shown at left; ~100 kDalton, highly basic proteins which contain two common domains, namely PIWI perhaps the RNase H enzyme, and PAZ consisting of 130 amino acids), to select and hold messenger RNA bearing a complementary sequence resulting in the cleavage of those messengers, preventing their translation into protein.
Small pieces of RNA also exercise control over gene expression by guiding the degradation of messenger RNA before it can be translated into protein by the cell's ribosomes. Several types of short RNAs provide powerful regulation of gene expression. The main classes of small RNAs are siRNAs (21 to 24 nucleotides long, so far identified in plant, fly, and worm cells, but also functional in mammalian cells), microRNAs (abbreviated miRNA), and PIWI-interacting, or piRNAs (24-31 nucleotides each, first isolated from reproductive cells of both flies and mannals), 21U RNAs (21 nucleotides long starting with uracil) and tncRNAs (noncoding RNAs from sections which don't code for proteins). These small RNAs are often cut off and separated from double-stranded RNA, the stem of an RNA hairpin which folds back on itself, or other sections of RNA which are not used as template for protein production. The small RNAs interacting with argonaute proteins (←shown at left; ~100 kDalton, highly basic proteins which contain two common domains, namely PIWI perhaps the RNase H enzyme, and PAZ consisting of 130 amino acids), to select and hold messenger RNA bearing a complementary sequence resulting in the cleavage of those messengers, preventing their translation into protein.
Both Linus Pauling and James Watson used a modeling procedure of creating models drawn to scale on paper to determine likely geometry and molecular fit. This is both a fast and easy way to help develop understanding of molecular structures. It is one than anyone with some understanding of chemical concepts can emulate.
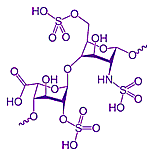
At right is heparin, a family of polymers composed of monosaccharides (sugar) which surround the outside of cells and interferes with the body's natural blood clotting mechanism. While it is not clear which molecules will lead to further advancements, this might be used to develop model building skills.
There are several conventions shown in this structural formula. None of the Carbon atoms are shown by letter (C). It is left for the viewer to understand that a Carbon atom occupies every unlabeled junction of bonds. In the two ring structures shown, each ring is composed of 5 Carbon atoms and 1 Oxygen atom. Carbon atoms have four valence electrons so nearly always forms four bonds to other atoms. Unless it has multiple bonds to another atom (such as the double bond to the Oxygen at the left side of this structure), This requires each Carbon to be bonded to four other atoms. If you count the bonds to each of the invisible
ring Carbons, you will note each apparently also has an invisible
bond and another invisible
atom. Those additional invisible
atoms are all Hydrogen. Generally the electron pairs forming chemical bonds stay as far apart as possible. This results in the four Carbon bonds maintaining an angle of separation of approximately 109.5° resulting in an arrangement with the attached atoms in the corners of a tetrahedron. Where bond are only in three directions (such as the Carbon double bonded to Oxygen on the left), the separation is approximately 120° placing the attached atoms in the corners of a flat triangle.
| atom | H | C | N | O | P | S |
| covalent radii (Å) | 0.32 | 0.72 | 0.68 | 0.68 | 1.04 | 1.02 |
| atomic radii (van der Waal, Å) |
1.10 | 1.55 | 1.40 | 1.35 | 1.88 | 1.81 |
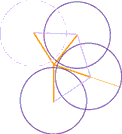
For example, the diagram at right was constructed as follows:
- Consider the bonds between atoms (drawn light purple) and the triangles formed when line segments are drawn to the center of the eventual ring (colored gold). If the bond angle between atoms is 109.5° (with half of that in each triangle), the angle between gold line segments must be 70.5°. That measure was used to draw each line segment.
- The circle for each atom was drawn centered on the end of each gold line segment with atomic radius a bit smaller than van der Waal's radius. Construction would be difficult if atoms were drawn larger, overlapping the center of the ring. Recall that the
edgeof each atom is actually vague as each atom's influence fades with distance.- One more than half the six ring atoms are drawn, with the faint extra (4th) to assist in construction alignment.
- Two of these patterns are printed on heavy paper and cut out around the outer perimeter.
- Each is folded, (your choice) either along the direction of each gold line or midway between lines and atoms. Folds are opposite so if one is folded inward, the adjacent folds are outward. If the extra construction atom is folded outward on one of the two sections, the other section should have it's extra folded inward (i.e., opposite).
- Each bold end atom is glued over the extra construction atom on the other section, aligning both the outer circle and the gold line segments. This should result in the puckered ring.
invisibleHydrogen atoms, print similar sections to those used to construct the ring. (The addition's atom with the light gold line is destined to become a duplicate representation of the ring atom.) Recall that both the sum of the covalent radii (gold line segment) and the atomic size (van der Waal's) should be smaller for an attached Hydrogen. (The Hydrogen nucleus should be inside the other atom!)
Communicating technical information such as observations and findings is a skill used by scientists but useful for most others. If you need course credit, use your observations in your journal to construct a formal report.
![]()
to next investigation
to Biochemistry menu
to e-Chemistry menu
to site menu